In Situ Hybridization Probe Design

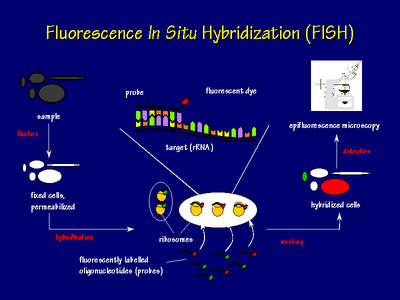
In situhybridization identifies where in the cellular environment a gene is expressed.
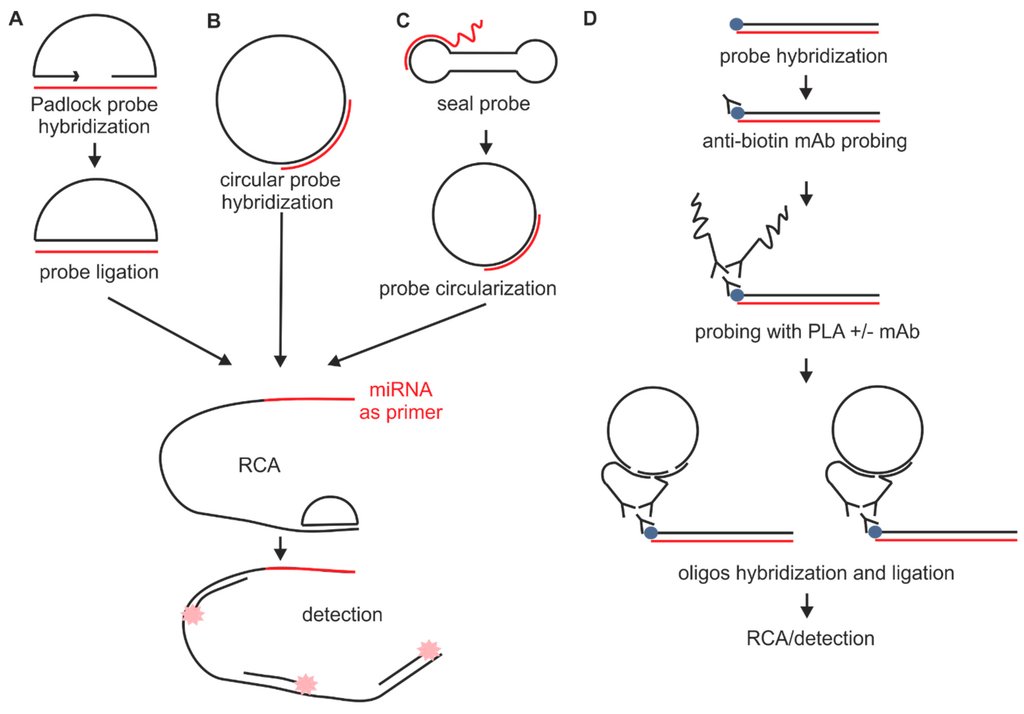
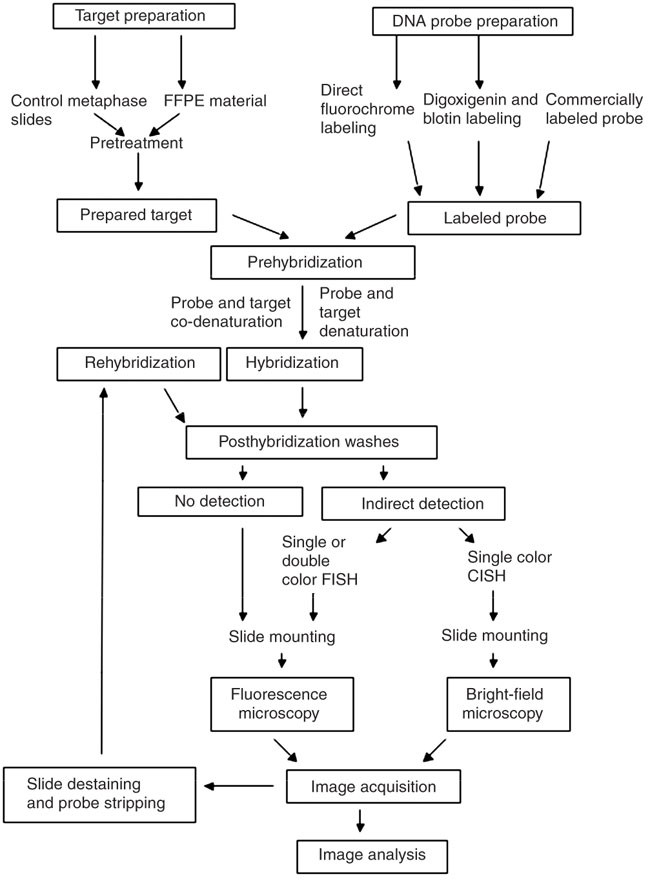
In situ hybridization probe design. Our rnascope in situ hybridization probe design pipeline can be applied to public or proprietary sequences for use with any of our chromogenic or fluorescent rnascope reagent kits for manual or automated assay configurations. Fixation and pre treatment of samples are followed by hybridization of a labeled probe by complementary base pairing to the target and. I prefer targeting the 3 end of the gene and try to anchor at least 1 2 or 2 3 of that probe s length to be in the 3 utr. The underlying basis of ish is that nucleic acids if preserved adequately within a histologic specimen can be detected through the application of a complementary strand of nucleic acid to which a reporter molecule is attached.
As far as designing in situ probes i typically ta clone 1 1 5kb probes. A labeled rna or dna probe can be used to hybridize to a known target mrna or dna sequence within a sample. In situ hybridization indicates the localization of gene expression in their cellular environment. In situ hybridization polynucleotide probe design software in situ hybridization of genes and mrna is most often based on polynucleotide probes.
Custom probe design hcr v3 0 probe sets enable multiplexed quantitative rna fluorescence in situ hybridization rna fish rna flow cytometry and northern blotting. Acd s made to order probe design process is highly flexible. In situ hybridization ish in situ hybridization ish is a unique molecular analysis method providing accurate localization of endogenous bacterial or viral nucleic acids such as dna mrna and microrna in metaphase spreads cells and tissue preparations. A labeled rna or dna probe hybridizes with a target mrna or dna sequence in a sample.
Probe design concept was still missing because the well established concept for oligonucleotide probe design cannot be transferred to polynucleotides. In situ hybridization ish is a technique that allows for precise localization of a specific segment of nucleic acid within a histologic section.